RMIT students developed an AI-powered solution to automate the transcription of medical documents.
Contributors
An Do, Nhat Tran, Quan Vu, Anh Nguyen, Phong Ho
Program
Bachelor of Information Technology
Notable achievements
Funded by RMIT Vietnam Strategic Innovation Challenge and featured at RMIT Experience Day to showcase RMIT’s contributions to innovation and healthcare
Background and motivation
Keeping medical records is crucial but often problematic, as doctors and staff spend a lot of time talking to patients and typing up details, which can lead to mistakes and impact patient safety. This administrative burden also detracts from direct patient care. We've identified this gap as an opportunity for technological intervention. We're developing software that captures live audio from nurses during documentation, using advanced speech recognition and natural lanquage processing to convert these audio notes into precise text that seamlessly integrates with existing medical systems.
The MedVoice project is driven by the need to improve the accuracy and efficiency of medical documentation in healthcare settings. Our primary objective is to develop a real-time audio-to-medical documentation solution that alleviates the burdens of manual transcription. This initiative is crucial in enabling healthcare professionals to focus more on patient care rather than administrative tasks. By allowing the medical staff to further interact with patient information with a AI ChatBot, MedVoice aims to be a universally adaptable tool that enhances the quality of patient care across diverse medical environments. The project’s evolution has been guided by the goal of creating a user-friendly, efficient system that meets the dynamic needs of modern healthcare.
System architecture
This project features a detailed system architecture, emphasizing the workflow for medical staff interaction and the utilization of the LLM pipeline for efficient audio data processing.
Workflow for medical staff interaction
The MedVoice application starts with the medical staff signing up and verifying their email via OTP (handled by Gmail SMTP Server). If successful, the app registers their account with the Backend. When the user decides to login; the system will check if the account exists or not. Once logged in, they are taken to the Medical Archive view with a first-time app tour. Users can record conversations with patients with live transcription displaying in-app, then click stop recording to send the recorugh the Medical Archive page. Additionally in the Medical Archive, a chat bot allows users to query patient information, with responses provided by the Backend based on the patient’s data.
LLMs pipeline
The LLMs pipeline begins with staff recording patient check-up audio, which is then sent to the Google Cloud socket. In the FastAPI backend, the audio recording is retrieved from the public domain. Next, it undergoes processing through the Whisper diarization model to separate speakers and generate corresponding transcripts. The results from Whisper diarization are fed into Meta Llama-70B. Prompt engineering ensures proper formatting of the data into JSON format, which is then posted back to the front-end of the system.
The background process of the MedVoice application contains two-step workflow within a healthcare processing system, starting with the transcription and structuring of patient audio data and culminating in its embedding and storage. In the first process, the LLM pipeline initiates by processing the recording through the Whisper-diarization model to distinguish speakers and generate a transcript. This transcript is then passed via LangChain to Meta's Llama3-70b-instruct model, which extracts and organizes relevant patient data into a structured JSON format (Patient Data). The Worker subsequently marks the task as DONE and sends the formatted medical transcript to the MedVoice Mobile App.
Concurrently, the second process is triggered, where the Patient Data is sent to an embedding model (ollama/msc-embed-text) for embedding and storage in a Vector Database (pgvector), ensuring efficient data retrieval for future queries. Importantly, these embedded data are personalized to the medical staff responsible for that patient, ensuring that they remain secure while maintaining efficient and secure interactions.
Experiments and resutls
This project conducted two experiments, focusing on LLM pipeline for recording and enhancing RAG system chatbot accuracy in medical data processing.
Experiment 1: LLM pipeline for recording
Datasets
- Medical Consultation Audio Recordings: A diverse set of patient-doctor interaction recordings, varying in accent, background noise, and medical terminology complexity.
- Transcribed and Annotated Data: Different JSON Schemas for practical medical documents.
Evaluation Metrics
- Word Error Rate (WER): Measures the accuracy of the transcription by comparing the model’s output against the manually transcribed data.
- Processing Latency: Measures the time taken from audio input to the generation of the final JSON output, aiming to optimize for real-time or near-real-time performance.
Base Models
- Whisper Model: Used for initial diarization and transcription of audio data.
- Meta Llama3-70B: Used for transforming the transcriptions into structured JSON format via prompt engineering and language understanding.
- LangChain: Orchestrating the pipeline and integrating the transcription and structuring steps.
Experiment 2: RAG system chatbot
Objective
Enhance the responsiveness and accuracy of the Retrieval-Augmented Generation (RAG) system in answering medical queries based on embedded patient data.
Datasets
- Embedded Patient Data: Embeddings generated from structured patient data, stored in the vector database, to be used as the knowledge base for the chatbot.
- Conversation Logs: Historical logs of queries and responses stored in the system to fine-tune the model on common and repetitive questions.
Evaluation Metrics
- Answer Accuracy: Measures the correctness of the responses generated by the chatbot when compared to the ground truth in the medical question-answer pairs.
- Embedding Retrieval Quality: Assesses the relevance and quality of the embeddings retrieved in response to a query, based on cosine similarity or another suitable distance metric.
- Prompt Injection Resilience: Evaluates the system's robustness against attempts to inject irrelevant or harmful prompts, ensuring that responses stay within the intended context and content. This metric is crucial for maintaining the integrity and safety of the responses, especially in a medical context.
Base Models
- Meta Llama3-70B: Utilized for understanding and generating responses based on retrieved embeddings and query context.
- ollama/nomic-embed-text: Used for generating embeddings of patient data and query text, which are then stored and retrieved from the pgvector database.
- Vector Database (pgvector): Responsible for storing embeddings and enabling efficient retrieval during query processing.
Design prototypes
 Onboarding screen of MedVoice with options to get started or log in.
Onboarding screen of MedVoice with options to get started or log in.
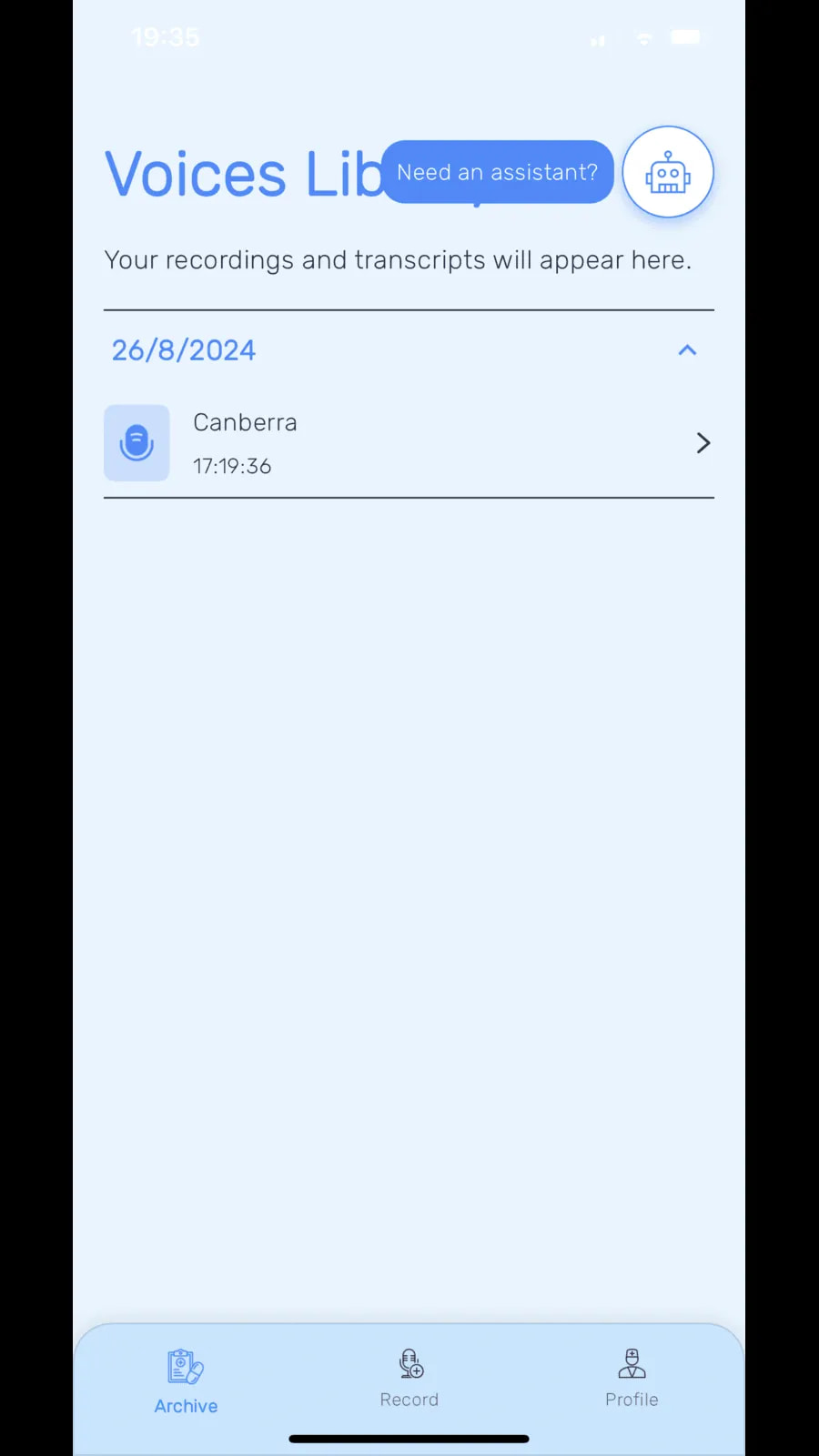 Voices Library displays saved recordings and transcripts, with a navigation bar offering Archive, Record and Profile functions.
Voices Library displays saved recordings and transcripts, with a navigation bar offering Archive, Record and Profile functions.
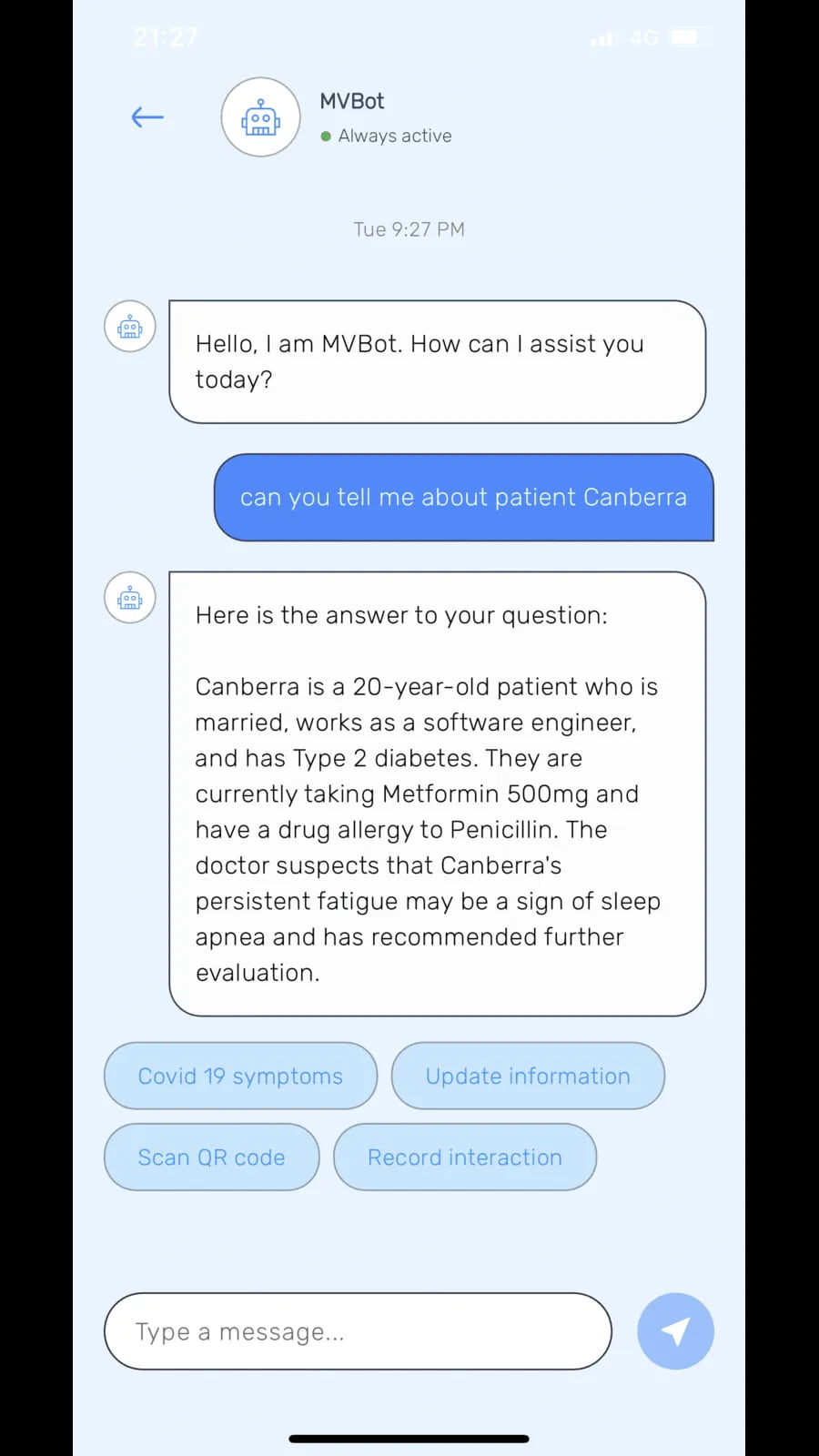 Chat interface with MVBot provides patient details and options like symptom updates, QR code scanning and recording interactions.
Chat interface with MVBot provides patient details and options like symptom updates, QR code scanning and recording interactions.


